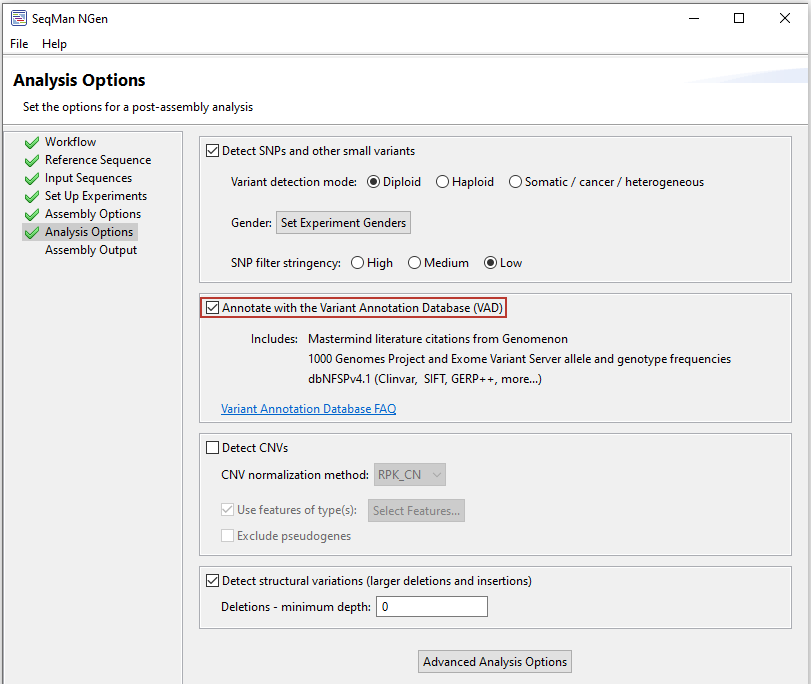
Lasergene Genomics automatically enriches the analysis of variants detected in your human resequencing data using our custom Variant Annotation Database, which integrates information from many large variant databases, including Mastermind, dbNFSP, the 1000 Genomes Project, and the Exome Sequencing Project, into your variant report. These databases provide a wide range of relevant data, including literature citation counts, predictions for disease potential, clinical significance reporting, evolutionary conservation scores, human genotype and allele frequencies in 25 human populations and impact on protein.
This integration is part of the automated pipeline offered by Lasergene Genomics which removes the complex, extensive manual intervention often required for detecting, annotating, and analyzing variants in human sequencing data.
Incorporating the variant data from the Variant Annotation Database into your project is as simple as checking a box when you set up your sequencing project in SeqMan NGen. Following assembly, access to detailed, comprehensive data from integrated databases is available for each identified variant in ArrayStar.