NovaFold
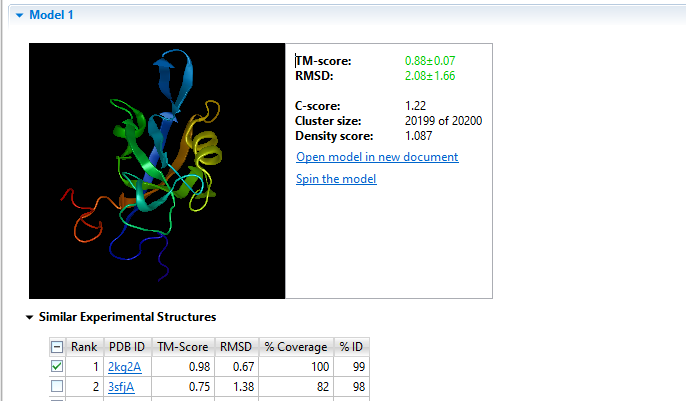
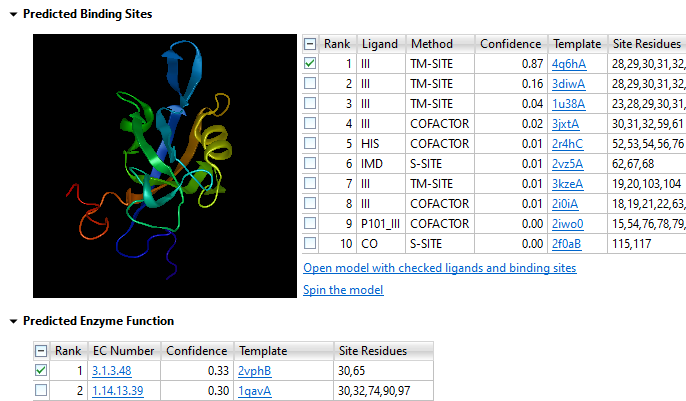
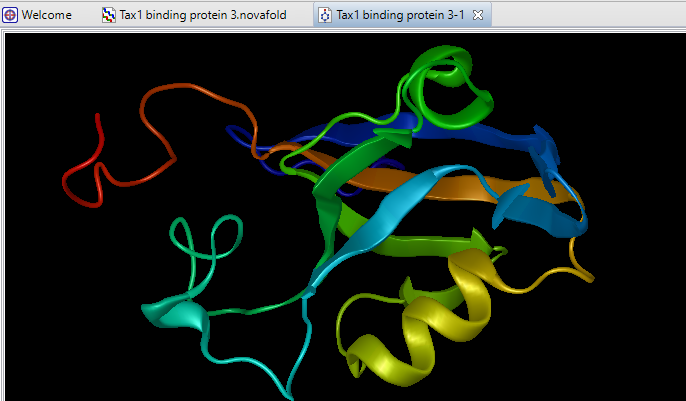
Determining the 3D structure of a protein is important for understanding its function, but doing this in a lab through x-ray crystallography is expensive and cumbersome, especially when a large number of protein sequences are involved. That’s why we developed NovaFold, our protein structure prediction software. NovaFold utilizes the international award-winning I-TASSER algorithms developed by Professor Yang Zhang’s laboratory at the University of Michigan that combine threading and ab initio folding technologies to build accurate, full 3D atomic models of proteins with previously unknown structures. View and manipulate models of each predicted structure in Protean 3D, where you can effortlessly analyze structural data, predicted binding sites, and protein function.
Resources
Please see our resources below for more information on NovaFold protein structure prediction software.
Protein Structure Prediction Workflow
How Can DNASTAR’s Protein Tools Help You?
Why Structure Prediction Matters
Nova Applications on Mac: Bringing the Power of Protein Modeling to the Desktop
Modeling GPCR Structures in NovaFold
Approaches to Protein Structure Prediction
Accuracy of NovaFold Protein Folding Predictions
High Resolution Protein and Antibody Modeling with NovaFold and NovaFold Antibody
NovaFold and I-TASSER: A winning combination for protein structure prediction and analysis
NovaFold Help
Quality models and useful insights
“Using the suite of DNASTAR programs, NovaFold, NovaFold Antibody, and NovaDock, I have generated functional models of antibody-antigen complexes from the component’s amino acid sequences. The quality of the models and the useful insights they generate more than justify the effort and computing time.”
FAQs
Do I need a Lasergene license to run NovaFold protein structure prediction software?
Yes. A Lasergene Protein license is required to run NovaFold. Protean 3D, included in Lasergene Protein, provides the interface for running NovaFold predictions and analyzing the results.
Do you offer a free trial of NovaFold?
One prediction for each of our Nova Applications is included in our Lasergene free trial. We also offer free, no-obligation, personalized demos of any of our Nova Applications with one of our scientists on staff, tailored to your specific research objectives.
What protein folding prediction method does NovaFold use?
NovaFold uses the I-TASSER algorithm developed by Professor Yang Zhang of the Department of Computational Medicine and Bioinformatics at the University of Michigan. This protein structure prediction…
NovaFold uses the I-TASSER algorithm developed by Professor Yang Zhang of the Department of Computational Medicine and Bioinformatics at the University of Michigan. This protein structure prediction algorithm utilizes a combination of threading and ab initio folding in predicting protein structure.
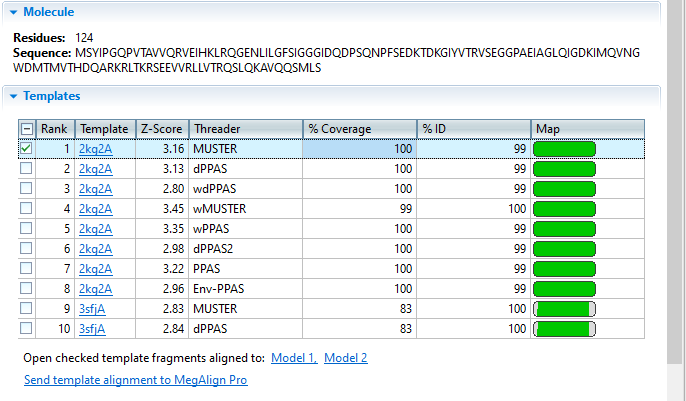
Threading attempts to match portions of the query sequence to template sequences. The template sequences, and their experimentally solved structures, are part of the RCSB Protein Data Bank (PDB). Ab initio folding uses biophysical properties of the query sequence and simulations to determine the likely structure(s) of the protein.